Research topics
Ecology and spillover of Nipah virus
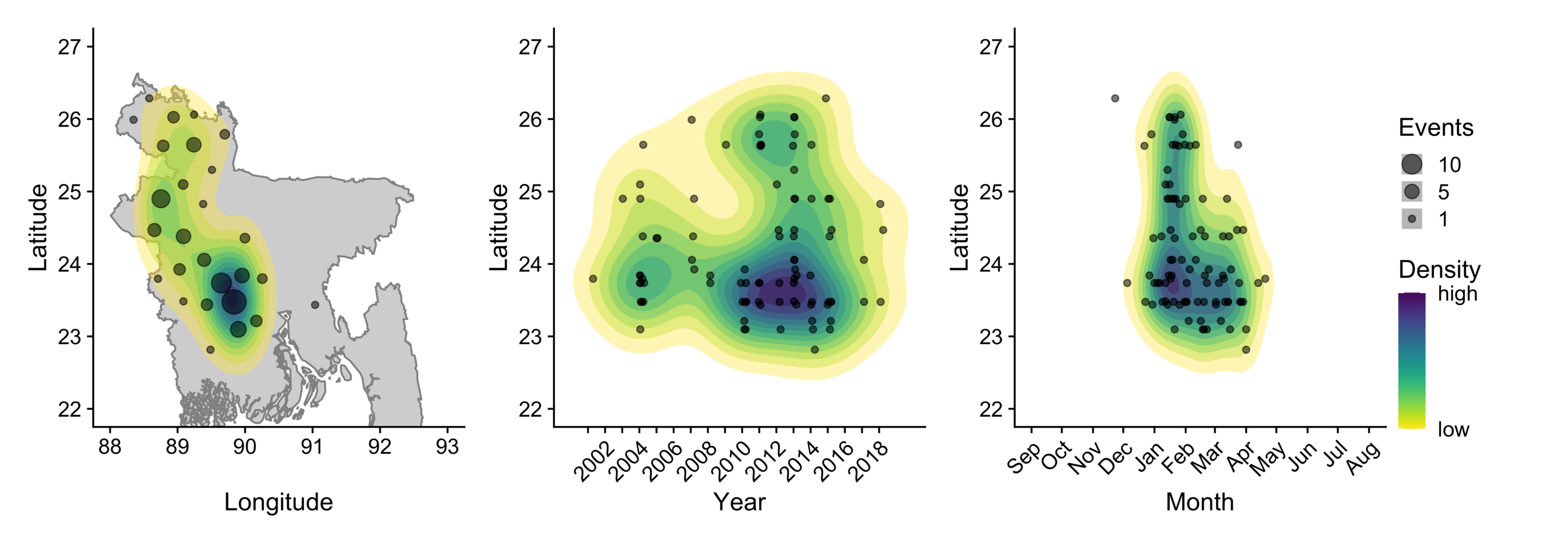
Nipah virus is a bat-borne infection that causes yearly outbreaks in Bangladesh with mortality >70% in human cases. I have contributed my expertise in bat biology and ecology to understand how human land use and animal behavior contribute to the emergence of Nipah virus at the human-animal interface. These insights are important for understanding how Nipah virus is maintained in bat populations and the seasonality of cross-species transmission to inform surveillance and prevention efforts.
- Cortes-Azuero et al. 2024 J Infect Dis
- McKee et al. 2022 Emerg Infect Dis
- Islam et al. 2021 EcoHealth
- McKee et al. 2021 Viruses
Phylogenetic comparative methods for assessing cross-species transmission
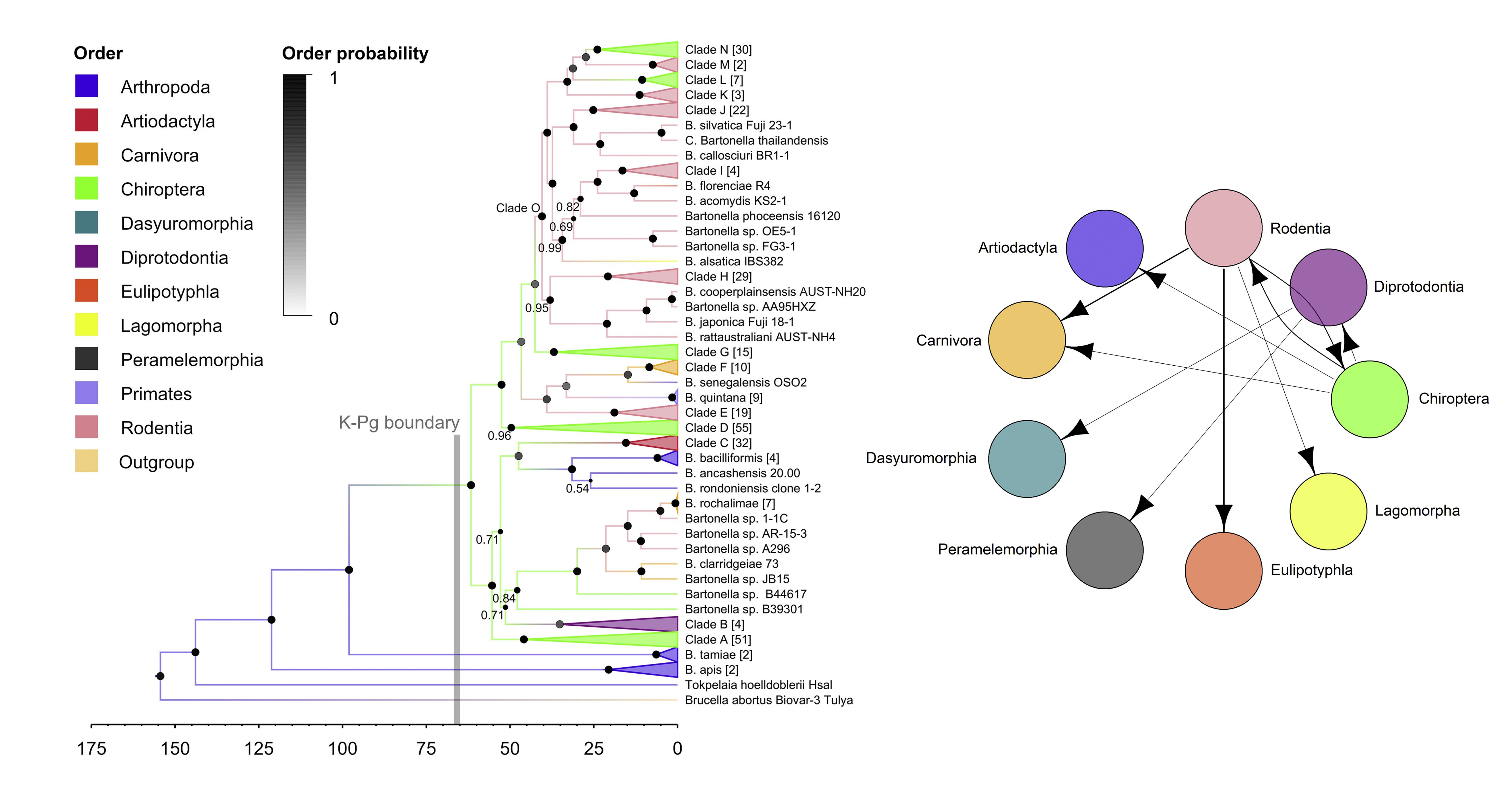
Pathogens frequently transmit between multiple closely related species in nature, but these processes are often unobserved due to lack of surveillance. Understanding the frequency and timing of these transmission events often has to been inferred from pathogen sequence data. Across several projects, I have used a combination of Bayesian phylogenetics, cophylogenetic approaches, and network analysis to understand how cross-species transmission of wildlife pathogens is structured by geography and host relatedness.
Population genetics and pathogen dynamics in wildlife
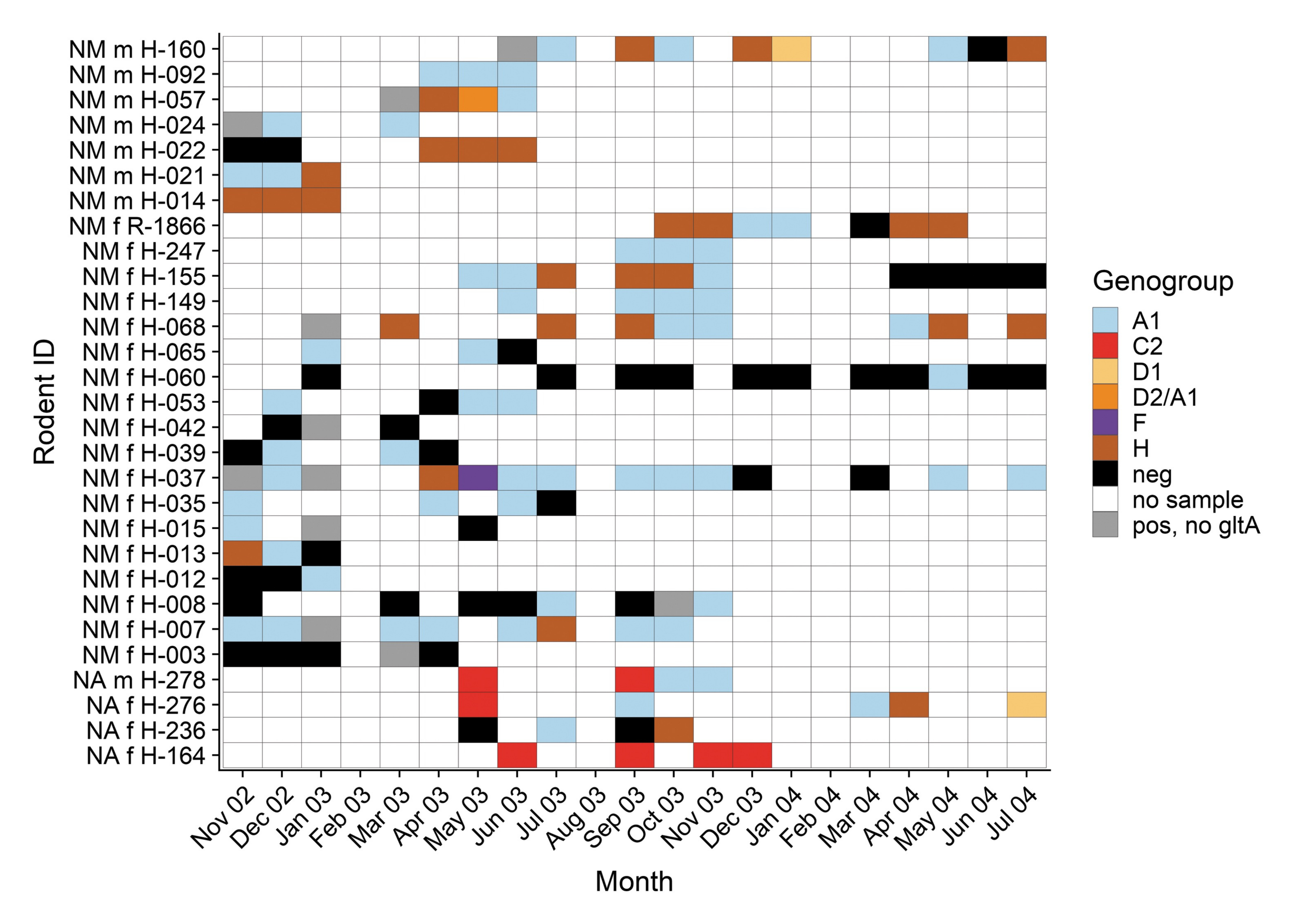
Longitudinal and spatially replicated surveillance studies are important to understand the geographic and temporal factors that contribute to disease dynamics and pathogen diversification in wild hosts. These types of studies can help to ascertain how pathogen transmission is structured across host populations, the seasonality of transmission, and how particular demographic groups are contributing to transmission.
- McKee et al. 2024 Parasitology
- Fagre et al. 2023 Microb Ecol
- Goodrich et al. 2020 PLOS ONE
- Goodrich et al. 2020 Vector Borne Zoonotic Dis
Molecular characterization of wildlife pathogens
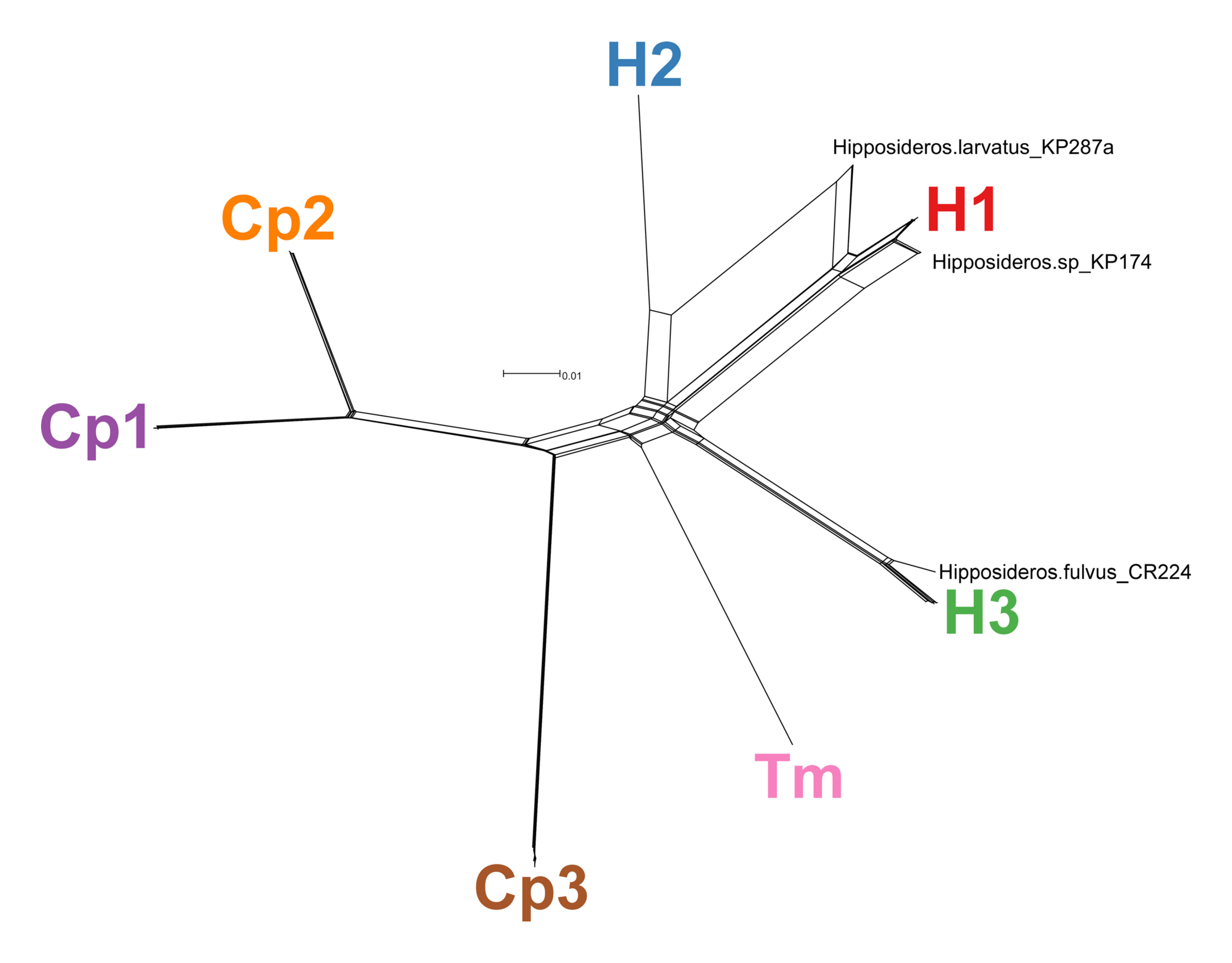
During my fellowship at CDC Division of Vector-Borne Diseases and beyond, I have contributed to the description of novel pathogens in wildlife. The bacterial genera Bartonella, Borrelia, and Leptospira are known to cause human disease, frequently following cross-species transmission from animals. It is important to understand the diversity of these bacteria and the processes that contribute to pathogen genetic diversity in nature, including mutation and recombination. This work involved sequencing of multiple genetic loci from cultured and uncultured bacteria and using various phylogenetic tools to understand the genetic diversity of these organisms and their relationships to known human pathogens.